Frontiers |
您所在的位置:网站首页 › imagej for mac › Frontiers |
Frontiers
ORIGINAL RESEARCH article
Front. Oncol., 16 August 2022Sec. Genitourinary Oncology
Volume 12 - 2022 |
https://doi.org/10.3389/fonc.2022.882888
ASNS can predict the poor prognosis of clear cell renal cell carcinoma Xinqiang Gan1,2†, Xinqiang Gan1,2†,  Ruiji Liu2†, Ruiji Liu2†,  Hong Cheng2*, Hong Cheng2*,  Weipu Mao1*, Weipu Mao1*,  Ninghan Feng3* and Ninghan Feng3* and  Ming Chen2,4*1Department of Urology, People’s Hospital of Putuo District, Shanghai, China2Department of Urology, Affiliated Zhongda Hospital of Southeast University, Nanjing, China3Department of Urology, Wuxi No.2 Hospital, Nanjing Medical University, Wuxi, China4Nanjing Lishui District People’s Hospital, Zhongda Hospital Lishui Branch, Southeast University, Nanjing, China Ming Chen2,4*1Department of Urology, People’s Hospital of Putuo District, Shanghai, China2Department of Urology, Affiliated Zhongda Hospital of Southeast University, Nanjing, China3Department of Urology, Wuxi No.2 Hospital, Nanjing Medical University, Wuxi, China4Nanjing Lishui District People’s Hospital, Zhongda Hospital Lishui Branch, Southeast University, Nanjing, ChinaPurpose: Clear cell renal cell carcinoma (ccRCC) is one of the most common malignancies of the urinary system. This study was conducted to discover a new target that can predict the prognosis and promote the treatment of ccRCC. Methods: The raw data were downloaded from the TCGA database, and the predictive value of ASNS for various clinicopathological features was verified in the following analysis. Then, we analyzed the potential involvement of ASNS in tumor immunity and obtained the possible pathways involving ASNS through GO/KEGG enrichment analysis and GSEA. We also further verified our findings in pathological specimens of ccRCC patients. Results: ASNS expression was significantly increased in ccRCC, which was associated with advanced clinicopathological characteristics. It was an independent prognostic factor for overall survival in 535 patients with ccRCC. Immune cell infiltration analysis revealed that ASNS expression was related to T lymphocyte infiltration of tumors and poor prognosis. Moreover, we performed relevant functional enrichment analyses of ASNS. Conclusions: ASNS might play a significant role in the development and immune cell infiltration of ccRCC and serve as a valuable clinical prognostic biomarker. IntroductionRenal cell carcinoma (RCC) is a common cancer type, and approximately 430,000 new global cases and 170,000 RCC-related deaths occurred in 2020 (1). RCC accounts for approximately 3% of all cancers, with the highest incidence in Western countries, and 80%–90% of RCC are clear cell renal cell carcinoma (ccRCC) (2). None of the treatments were effective in patients with renal tumors subjected to the same surgical procedures and patients with advanced disease treated with similar drugs (3). Although an occasional response was reported, the available systemic therapies did not increase the survival of patients with advanced disease (2). Early detection and screening are priorities for RCC research (4). Therefore, discovering a new target of ccRCC, especially for those with advanced and metastatic diseases, is crucial. Asparagine synthase (ASNS) catalyzes the synthesis of asparagine and glutamate from aspartic acid and glutamine in an ATP-dependent amidotransferase reaction, accompanied by glutamine deamidation (5). Large-scale loss-of-function analysis in vitro identified ASNS as cancer dependent in several solid malignancies; however, the specific mechanism has not been discovered (6). Knott et al. highlighted the role of ASNS in tumor growth and metastatic dissemination in a breast cancer model (7), prompting the necessity to evaluate the expression of ASNS and facilitating the prognosis of patients with ccRCC. We downloaded raw data from The Cancer Genome Atlas (TCGA) database and verified the predictive value of ASNS for various clinicopathological features in the following analyses. We analyzed the potential involvement of ASNS in tumor immunity and identified the possible pathways associated with ASNS through Gene Ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses and gene set enrichment analysis (GSEA). We further verified our findings in the pathological specimens of patients with ccRCC. To conclude, we found that ASNS was highly expressed in ccRCC patients, and its high expression could lead to a worse prognosis. Thus, our findings revealed that ASNS might play a significant role in the development and immune cell infiltration of ccRCC and serve as a valuable clinical prognostic biomarker. Materials and methodsPublic database collectionGene expression (535 tumor and 72 normal samples), DNA methylation, phenotype, and survival data were downloaded from http://xena.ucsc.edu/. |logFC| > 1.5 and p < 0.01 were defined as the criteria for differentially expressed genes (DEGs), and DEGs were identified using the limma R package. COX univariate and multivariate analysisCox univariate and multivariate analyses were performed to determine the risk factors for ccRCC prognosis using R software. Survival analysisOverall survival (OS) and progression-free survival rates of patients with ccRCC were investigated by Kaplan–Meier analysis. Correlation between immune infiltration and expressionTIMER2.0 (http://timer.comp-genomics.org/) was used for the comprehensive analysis of the relationship between ASNS expression and tumor-infiltrating immune cell levels, namely, CD4+ T cells, Tregs, CD8+ T cells, CTLA4, and PD-L1 (CD274) (8). Expression of hub genes and survival analysisThe STRING (https://string-db.org/) website analyzes protein–protein interactions using a unique set of computer prediction models (9). The expression levels of hub genes in tumor and normal tissues based on the ccRCC dataset were compared using the Wilcoxon rank-sum test (p < 0.05). OS analysis for the expression of hub genes between the high- and low-expression groups was performed, with a p-value of 0.5, p-value < 0.05). The Clusterprofiler R package was used for GO enrichment analysis, which included molecular function, biological processes, and cellular components of DEGs, and KEGG enrichment analysis. Gene set enrichment analysisThe molecular mechanisms involved in ccRCC with different levels of ASNS expression were examined by the GSEA approach (11). The reference gene set used for GSEA was obtained from c7.immunesigdb.v7.5.1.symbols.gmt. Gene sets with a nominal p-value less than 0.05, |Normalized Enrichment Score| greater than 1, and false discovery rate less than 0.25 in the GSEA report were considered statistically significant. Western blottingWestern blotting was performed as described in our previous study (12). ImmunohistochemistryThe expression and distribution of ASNS protein were detected by immunohistochemistry in paraffin-embedded tissue sections of each group. After routine paraffin dewaxing to water, antigen repair was performed. Hydrogen peroxide solution (3%) was added to remove endogenous catalase. Bovine serum albumin sealing solution (1%) was added, and the slides were incubated for 15 min. After the blocking solution was added, the primary antibody against ASNS (1:100 dilution) was added, and the slides were incubated overnight at 4°C. Thereafter, they were washed thrice with phosphate-buffered saline with Tween 20. The secondary antibody (1:100 dilution) was added, and the slides were incubated for 1 h at room temperature. DAB was added for color development, after which the slides were re-dyed with hematoxylin for 30 s. After washing with running water for 1 min, the slides were treated with 0.1% sodium bicarbonate to develop blue color. Then, the slides were subjected to dehydration and xylene treatment until the sections became transparent, mounted with neutral gum, observed under a microscope, and photographed. The immunohistochemical staining intensity of ASNS was assessed using ImageJ software with the assistance of experienced pathologists in Affiliated Zhongda Hospital of Southeast University (13, 14). Sample informationClinical data of 81 patients were collected at the Affiliated Zhongda Hospital of Southeast University from March 2019 to May 2022. All patients were diagnosed with ccRCC (unilateral) and had no other carcinomas. All patients did not undergo any preoperative therapy for the carcinoma (chemotherapy, targeted therapy, immunotherapy, etc.). Age distribution: 19–85 years old. Surgical modalities: Partial nephrectomy or radical nephrectomy. Neoplasm histological grade criteria: Fuhrman nuclear grading system. Pathological stage criteria: American Joint Committee on Cancer (AJCC). Statistical analysisAnalyses were performed using IBM SPSS for MAC version 26.0. The Mann–Whitney test was used to compare continuous variables between the two groups. The immunohistochemical staining intensity and clinical characteristics, such as gender, age, location, tumor size, TNM stage, AJCC stage, and grade were subjected to Spearman rank correlation analysis. p-values less than 0.05 were considered statistically significant. ResultsOverexpression of ASNS mRNA in ccRCC is related to poor prognosisThe expression pattern of ASNS in multiple pan-cancers was evaluated by using data from TIMER2.0, and the prognostic values of ASNS in multiple cancers (invasive breast carcinoma, cervical squamous cell carcinoma, endocervical adenocarcinoma, cholangiocarcinoma, esophageal carcinoma, kidney renal clear cell carcinoma, hepatocellular carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, pancreatic adenocarcinoma, and stomach adenocarcinoma) are shown in the survival map prepared using GEPIA2 (Figures 1A, B). The pan-cancer overexpression of ASNS was linked to poor prognosis. The mRNA data of ccRCC were downloaded from TCGA (https://xenabrowser.net/datapages/) and included 535 tumor tissues and 72 normal tissues. ASNS was highly expressed in ccRCC compared with normal renal tissues (p < 0.001; Figure 1C) and was correlated with poor prognosis, i.e., OS (p < 0.001; Figure 1D). Furthermore, the high protein expression level of ASNS in ccRCC was verified by data from the Human Protein Atlas (HPA) databases (https://www.proteinatlas.org/) (Figure 1E). FIGURE 1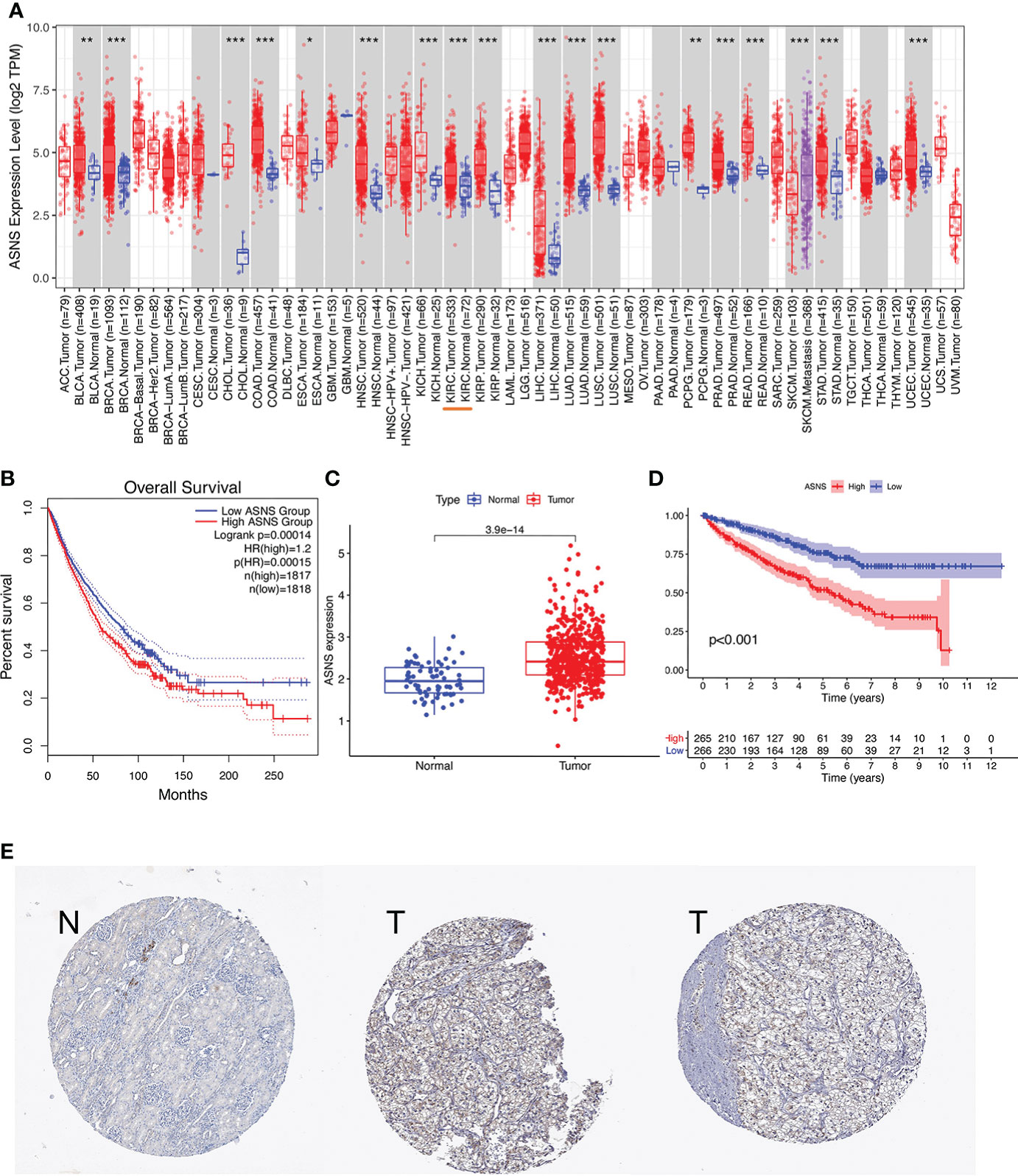 Figure 1 Expression of ASNS. (A) Pan-cancer expression of ASNS. (B) Prognostic values of ASNS gene in multiple cancers (BRCA, CESC, CHOL, ESCA, KIRC, LIHC, LUAD, LUSC, PAAD, and STAD). (C) ASNS expression in ccRCC and normal tissues. (D) Relationship between ASNS expression levels and prognosis, i.e., overall survival (OS). (E) ASNS protein level in ccRCC from the HPA database. The statistical significance computed by the Wilcoxon test is annotated by the number of stars (*: p-value 0.6; Figure 3C). Subsequently, serial ROC analysis showed favorable diagnostic values for ASNS to predict various clinicopathological features (Figures 3D–K). Ultimately, the above results were validated using nomogram and calibration plots, which showed that ASNS had excellent potential for predicting clinicopathological features in ccRCC (Figures 3L, M). FIGURE 3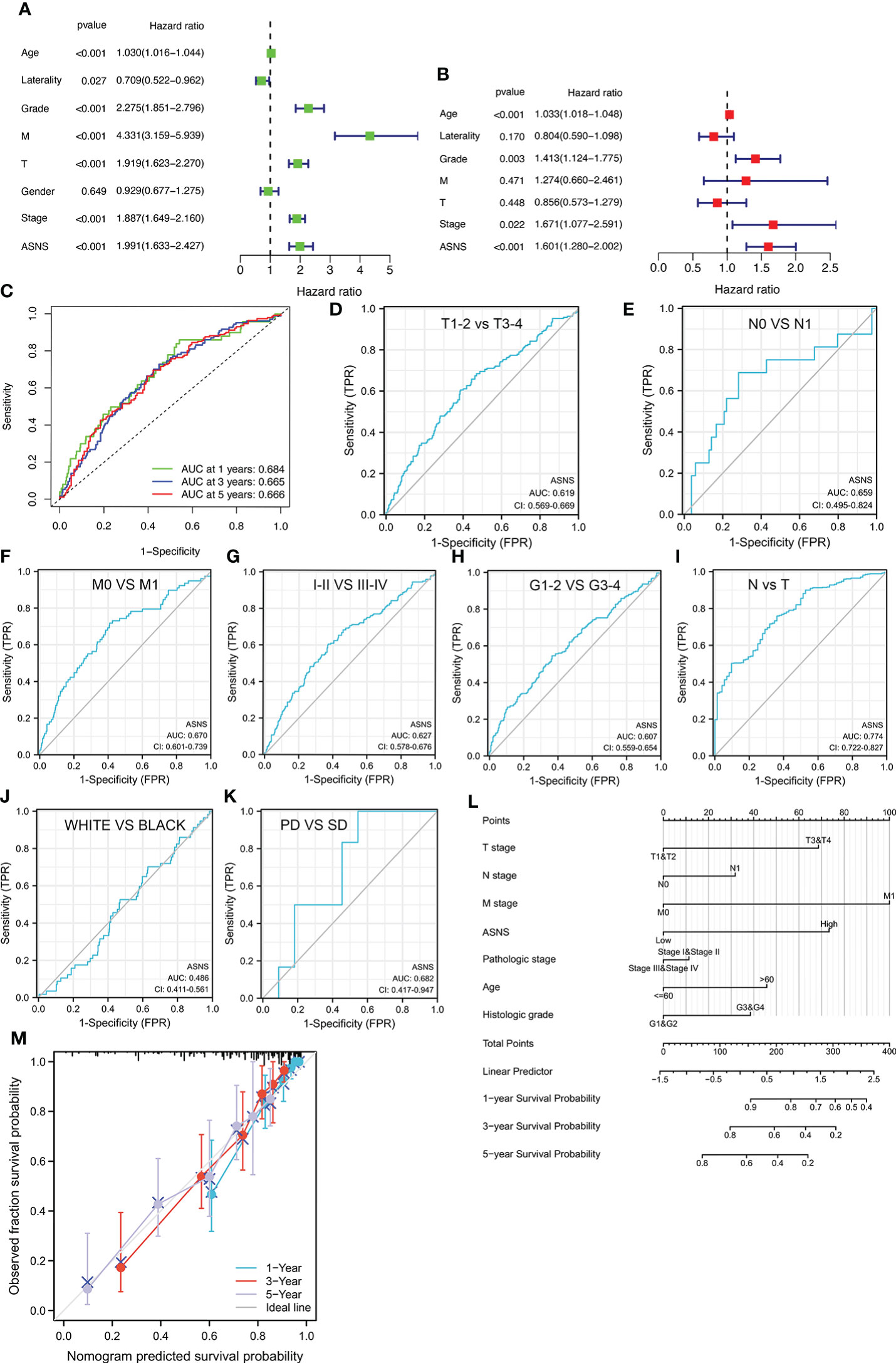 Figure 3 Identification of ASNS as an independent prognostic factor for ccRCC. (A, B) Univariate and multivariate independent prognostic analysis of ASNS. (C) Receiver operating characteristic (ROC) curve predicting the correlation between ASNS expression and OS. (D–K) ROC curve predicting the correlation between ASNS expression and clinicopathological features [T1–2 vs. T3–4, N0 vs. N1, M0 vs. M1, pathological stage I–II vs. III–IV, neoplasm histological grade 1–2 vs. 3–4, normal vs. tumor, race (white vs. black), and primary therapeutic outcome, i.e., partial disease vs. stable disease]. (L, M) Nomogram and calibration plots, respectively. DNA methylation analysis of ASNS geneTo further explore the regulation of ASNS in ccRCC, we performed a series of methylation analyses. Methylation levels were detected at the promoter region sites of the ASNS gene (Figure 4A), and ASNS expression was inversely proportional to the methylation level (Figures 4B, C). We further determined the relationship between ASNS gene methylation level and various clinicopathological features such as pathologic TNM stages (Figures 4D–F), tumor stages (Figure 4G), and neoplasm histologic grades (Figure 4H) of ccRCC. The results demonstrated that the methylation level of ASNS gradually decreased with tumor progression. Furthermore, the lower methylation level of ASNS led to a worse prognosis, such as poorer OS rates (p < 0.001; Figures 4I–K) and poorer progression-free survival (p < 0.001; Figure 4L). The decrease in the methylation level of ASNS with tumor progression indicates that the methylation modification of ASNS plays a key regulatory role in the expression of ASNS. FIGURE 4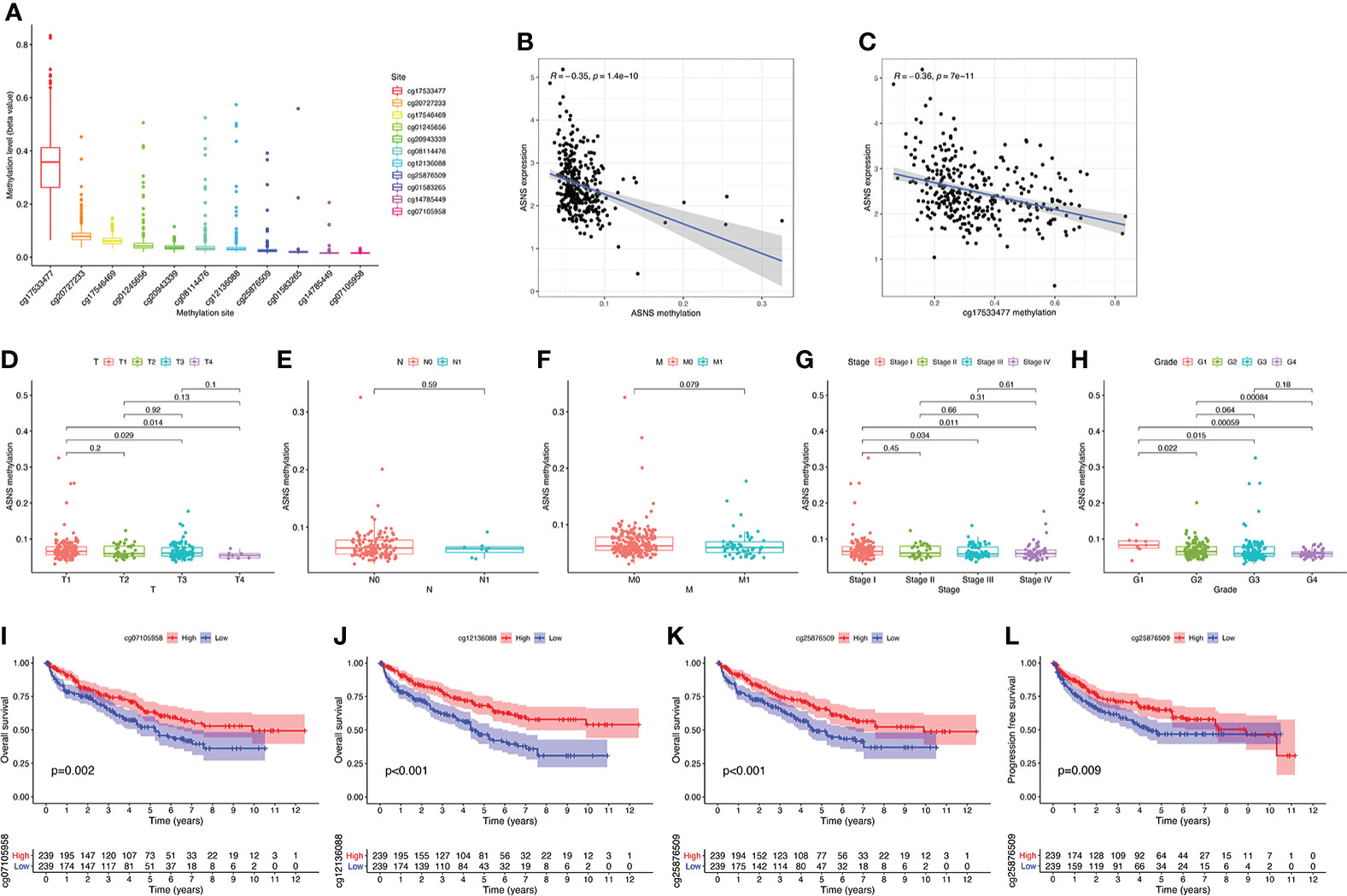 Figure 4 DNA methylation analysis of ASNS. (A) Methylation levels at different sites of ASNS. (B, C) The relationship between ASNS expression level and methylation level. (D–H) ASNS methylation in samples stratified by T stage, N stage, M stage, neoplasm histological grade, and pathological stage. (I–L) Methylation levels of ASNS on the prognosis of ccRCC (OS and progression-free survival rates). Immune infiltration analysis of ASNS in ccRCCccRCC has a relatively high stromal score and immune score among common cancers (15). High immune scores and infiltration of Tregs are significantly associated with poor OS, high tumor stage, and more chances of metastases in ccRCC (16). To further investigate the relationship between ASNS expression and the immune microenvironment in ccRCC, we performed immune infiltration analysis using TIMER2.0. The immune infiltration level of non-regulatory CD4+ T cells was negatively correlated with ASNS expression (Figure 5B), whereas Tregs were positively correlated (Figure 5C). Subsequent analyses showed that higher CD4+ T-cell infiltration was associated with better prognosis (Figure 5E), and higher Tregs infiltration was associated with poorer prognosis (Figure 5F). The correlation between CD8+ T-cell infiltration and ASNS was also analyzed; however, the results were insignificant (Figures 5A, D). Finally, ASNS expression correlated positively with the expression of immune checkpoints such as CTLA4, and PD-L1 (CD274) was positive (Figures 5G, H). FIGURE 5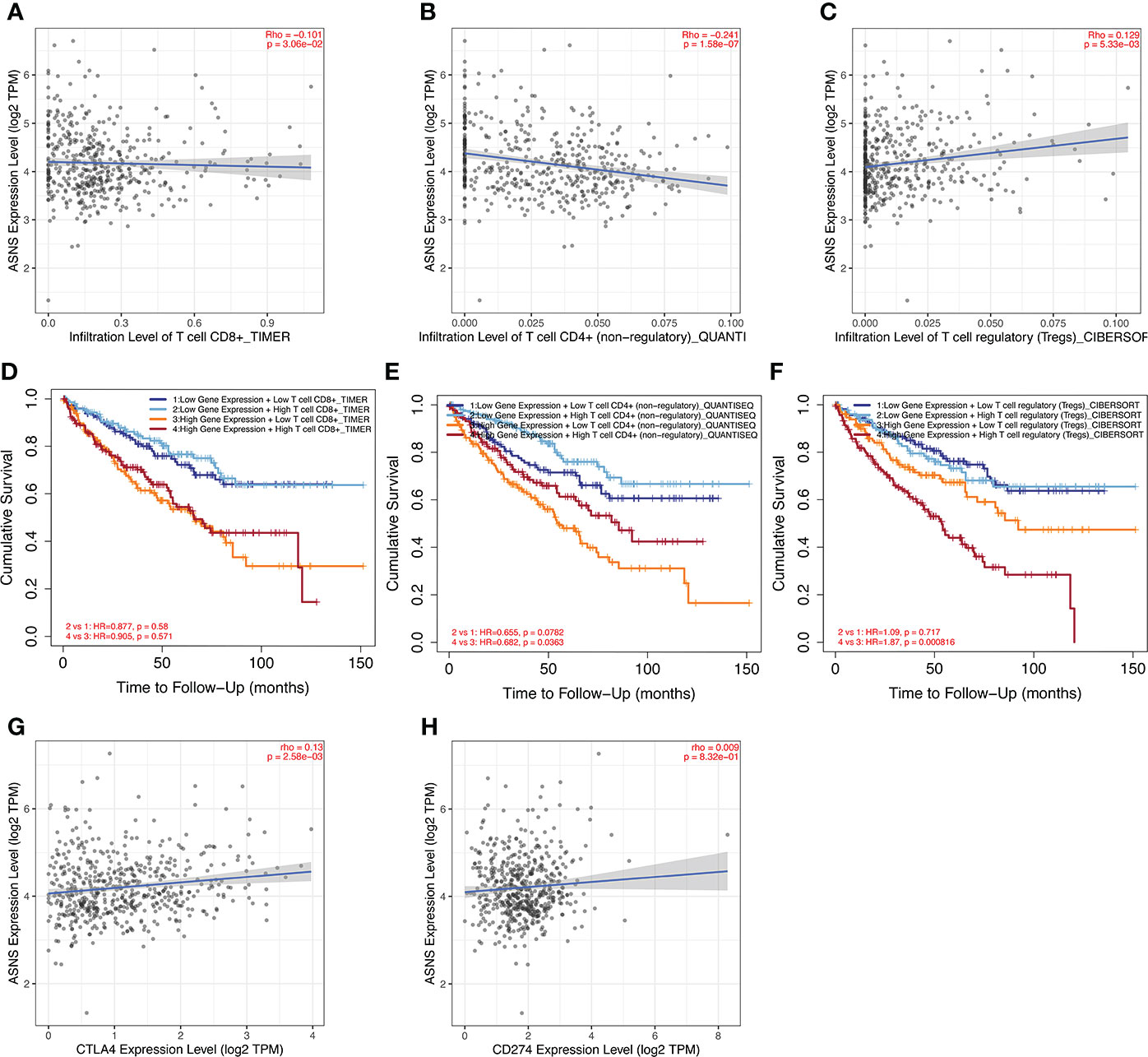 Figure 5 Immune infiltration analysis of ASNS in ccRCC. (A–C) Correlation between ASNS expression and immune cell infiltration in ccRCC. (D–F) Correlation between immune cell infiltration and prognosis. (G, H) Correlation between ASNS and expression level of immune checkpoint molecules. Cell function and pathway analysis in ASNSWe analyzed the positively and negatively correlated genes of ASNS in ccRCC and plotted heatmaps to show the expression patterns of the top 20 ASNS-associated genes (Figure 6A). To further determine the functions of ASNS in tumor progression and prognosis, GO and KEGG enrichment analyses were performed. GO enrichment analysis found that ASNS protein may be located on the cell membrane and regulate the molecular transport in cells (q-value |
【本文地址】